REGISTRATION – Version 1.0, released November 2024
The unique software that can identify and categorise different types of atoms within atomistics simulations or models, and to perform inter-atomic interaction analysis. This involves classify, annotate and quantity atomic interactions to provide statistical and structural information of a molecular system.
D_ATA contains two unique features implemented in DL_FIELD and DL_ANALYSER, respectively: (i) DL_F Notation to describe chemical characteristics of atoms and, (ii) DANAI, which is a chemical-based language construct to annotate non-bonded atomic interactions. Both features are complimentary to each other to create a universal atomic expression syntax that is independent of how the molecular systems are derived, either from molecular simulations or experimental measurements.
Functions
- Reads atomic configurations expressed in simple xyz and Gromacs gro formats.
- Identification and annotation of the chemical nature of constituent atoms in molecular systems (Typer). Works only for organic molecules.
- Identification and annotation of atomic and molecular non-bonded interactions in the system (Typer and Analyser) without the use of diagrammatic or pictorial illustration.
- Enabling atomic interactions to become data discoverable and searchable.
- Quantification of non-bonded interactions to rationalise the roles these interactions play in molecular systems (Analyser).
- Characterisation of non-bonded interactions in terms of geometrical orientations and shapes.
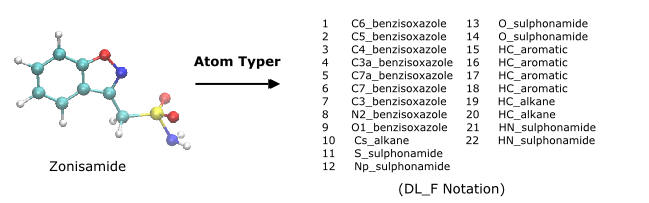
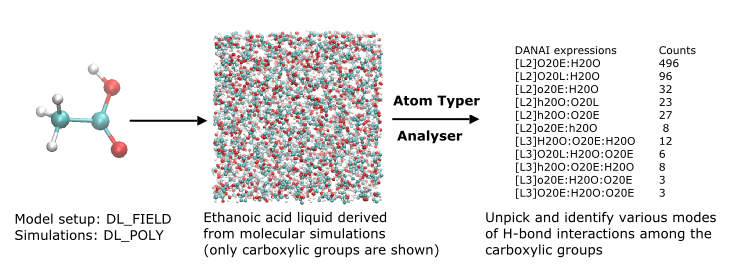
DL_F Notation:
- Journal reference: C.W. Yong, ‘Description and implementations of DL_F Notation: A natural chemical expression system of atom types for molecular simulations’, J. Chem. Inf. Model. 56, 1405-1409 (2016)
- Describes the universal expression atom typing as in force field setup for molecular simulations, available at SSRN: http://dx.doi.org/10.2139/ssrn.4254942
- More information, available as a tutorial for DL_FIELD, in DL_Software Digital Guide (DL_SDG):
DANAI:
- Journal reference: C.W. Yong, I.T. Todorov ‘DL_ANALYSER Notation for Atomic Interactions (DANAI): A natural annotation system for molecular interactions, using ethanoic acid liquid as a test case’ Molecules 23, 36 (2018)
- More information, available as tutorial for DL_ANALYSER, in DL_Software Digital Guide (DL_SDG): https://dl-sdg.github.io/RESOURCES/TUTORIALS/dla_8.html
Example cases:
- I. Rosbottom, et, al. ‘The integrated DL_POLY/DL_FIELD/DL_ANALYSER software platform for molecular dynamics simulations for exploration of the synthonic interactions in saturated benzoic acid/hexane solutions’, Mol. Sim. 47, 1 (2019)
- M. Guo, et. al. ‘Triglycine (GGG) Adopts a Polyproline II (pPII) Conformation in Its Hydrated Crystal Form: Revealing the Role of Water in Peptide Crystallization’, J. Phys. Chem. Lett. 12, 8416 (2021)
- I. Rosbottom, et. al. ‘The Structural Pathway from its Solvated Molecular State to the Solution Crystallisation of the α- and β-Polymorphic Forms of Para Amino Benzoic Acid’, Faraday Discussions 235 (2022)
- V.W. Barron, et. al. ‘Comparison between the Intermolecular Interactions in the Liquid and Solid forms of Propellant 1,1,1,2 – Tetrafluoroethane’, J. Mol. Liq. 383, 121993 (2023)
- C.W. Yong, et. al. ‘Data on the intermolecular interactions of 1,1,1,2-tetrafluoroethane liquids from molecular dynamics simulations, Data in Brief 50:109485 (2023)
